Source:Professor Chin-Fa Lee (Distinguished professor and Head of the Department of Chemistry at NCHU)
The clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (CRISPR/Cas9) system has been rapidly shifted from its natural role as an RNA-guided genetic adaptive immune system in prokaryotes to a robust site-specific gene editing method. The CRISPR/Cas9 system consists of two critical components which is the Cas9 endonuclease and a short single-guide RNA (sgRNA) which forms a Cas9•sgRNA ribonucleoprotein (RNP) complex. Based on a simple base-pairing mechanism, the RNP identifies and cuts a targeted DNA in the genome, which leads to a double-strand break (DSB) at the specified location. The Endogenous DNA repair mechanisms such as the non-homologous end joining (NHEJ) pathway, leads to insertions or deletions (indels) that results in gene disruption. Gene disruption via CRISPR/Cas9 editing has been frequently applied to knock down genes in cell lines and animal models. Alternatively, complete gene knockout can be achieved via CRISPR/Cas9-mediated gene deletion, where a pair of sgRNA targeting two ends of a given gene is used in the presence of Cas9 protein to induce two DSBs at the targeted sites where subsequent NHEJ-based DNA repair enables precise removal of the gene. In contrast to gene disruption, CRISPR/Cas9-mediated gene deletion offers a different gene knockdown solution capable of removing a DNA sequence (up to 30 million bp) associated with monogenetic diseases, like Duchenne muscular dystrophy (DMD) and Leber congenital amaurosis type 10 (LCA10).

Chin-Fa Lee, a Lifetime distinguished professor and Head of the Department of Chemistry at NCHU, collaborates with scientists from UCLA and recently published in “Small”, an authoritative journal in this field. This technology platform provides solutions for the clinical treatment of patients with genetic diseases of Duchenne muscular dystrophy (Duchenne muscular dystrophy) and also showed a greater gene deletion efficiency in the human cardiomyocyte cell line (AC16) induced pluripotent stem cells and mesenchymal stem cells.
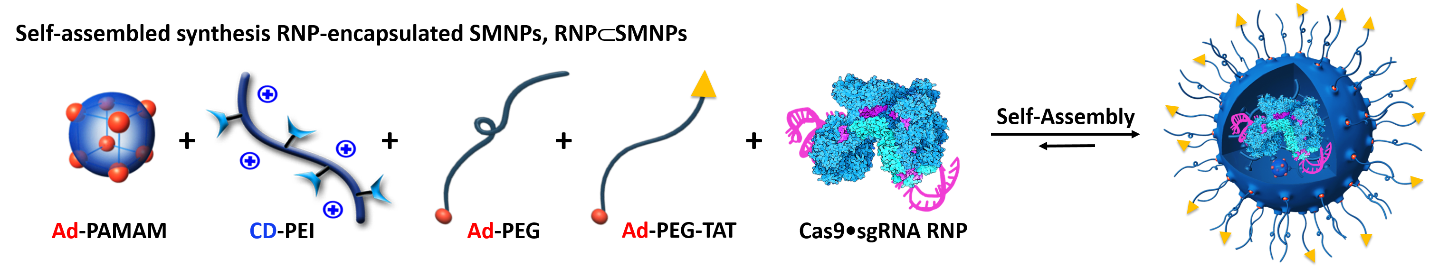
Self-assembled synthesis of RNP-SMNPs
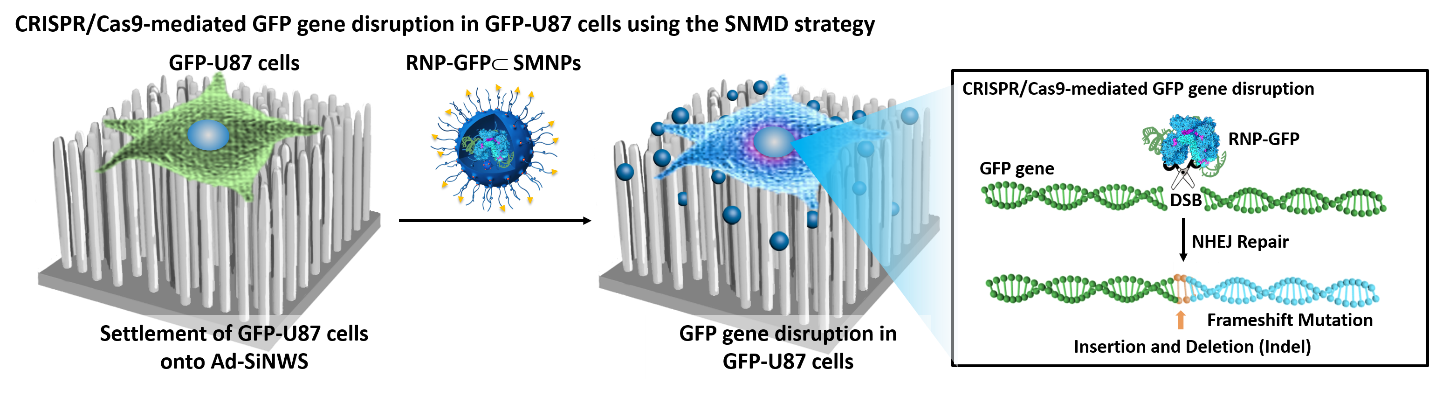
CRISPR/Cas9 mediated SNMD system for GFP-U87 as a model cell line
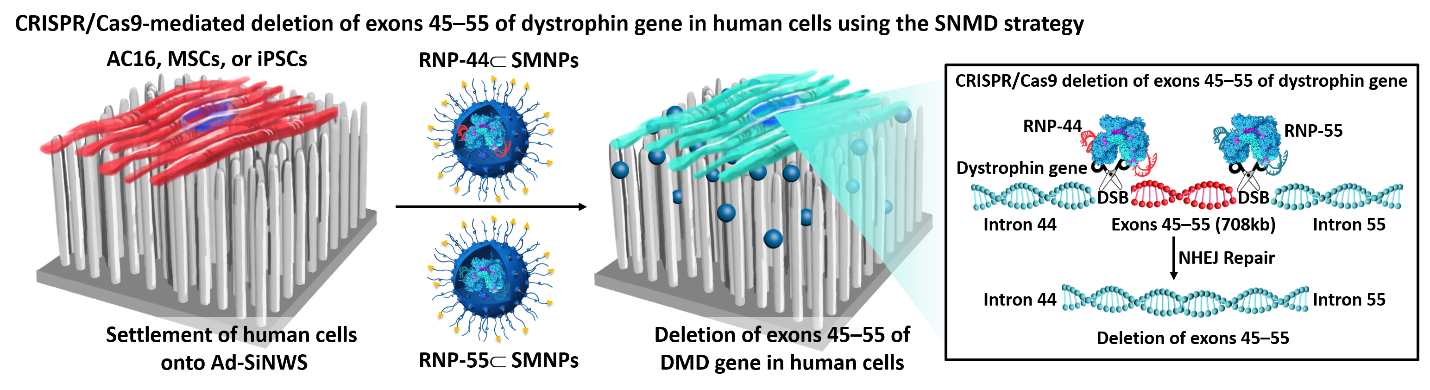
CRISPR/Cas9 mediated SNMD system for AC16, MSCs in human cell linesB
We have successfully demonstrated the feasibility of applying a SNMD-based strategy to deliver Cas9 RNP complexes into target cells, enabling CRISPR/Cas9-mediated gene disruption and deletion. This SNMD strategy leverages the local enrichment of RNP⊂SMNPs from surrounding medium onto nanowires of Ad-SiNWS. As cells come in contact with the nanostructured substrates, physical interactions between nanowires and cell membranes facilitates the uptake of RNP⊂SMNPs. In our previous work, we found that the potential of SNMD-based strategy is an effective delivery platform to perform HBB/ GFP knocking into the AAVS1 locus via homology-directed repair pathway in hematologic cells to cure hemoglobinopathies. Here, through co-delivering a pair of RNP complexes, we achieved high efficiency of large deletion of exons 45–55 (708 kb) in the dystrophin gene via NHEJ pathway in human adherent cells, including AC16 cells, MSCs, and iPSCs, offering a general clinical therapeutic solution for DMD. This is the longest gene deletion record using non-viral vectors in the literatures. Future studies will focus on validating the feasibility of this system to deliver CRISPR/Cas9 systems including RNP, DNA plasmids, and mRNA to correct mutated genes, resulting in broadening the types of diseases that could be targeted by our approach. To explore the potential in vivo applications of this platform, we are planning to develop an implantable nanosubstrate that is biodegradable and flexible (e.g., based on polymer nanoneedles) to enable implantation into target organs. This work is currently in progress.